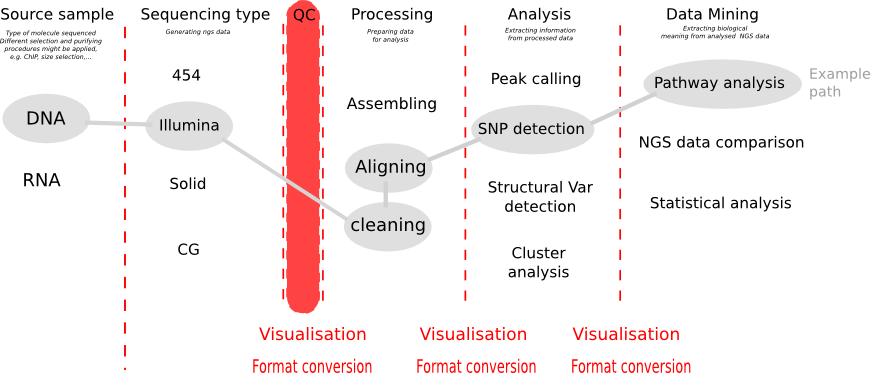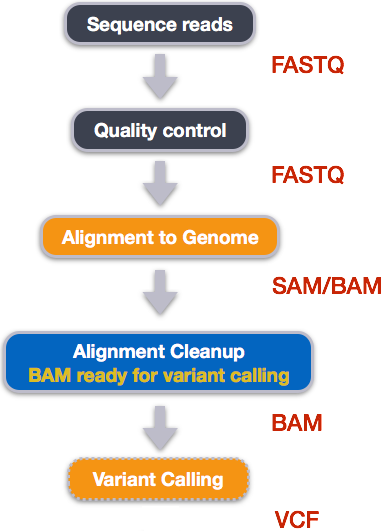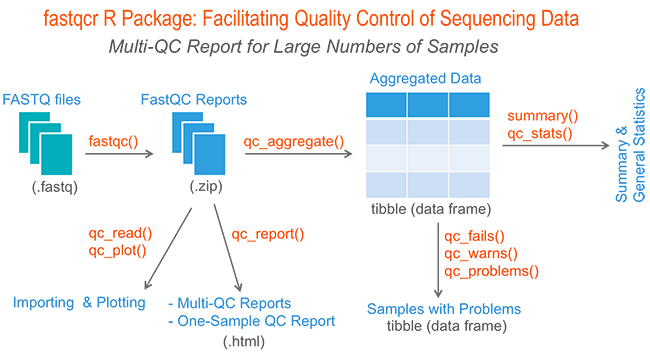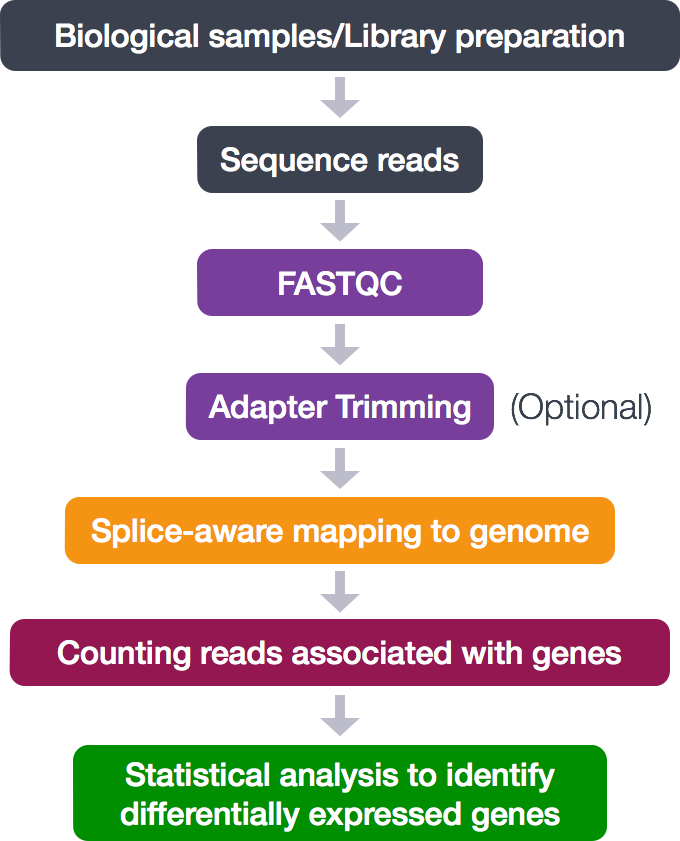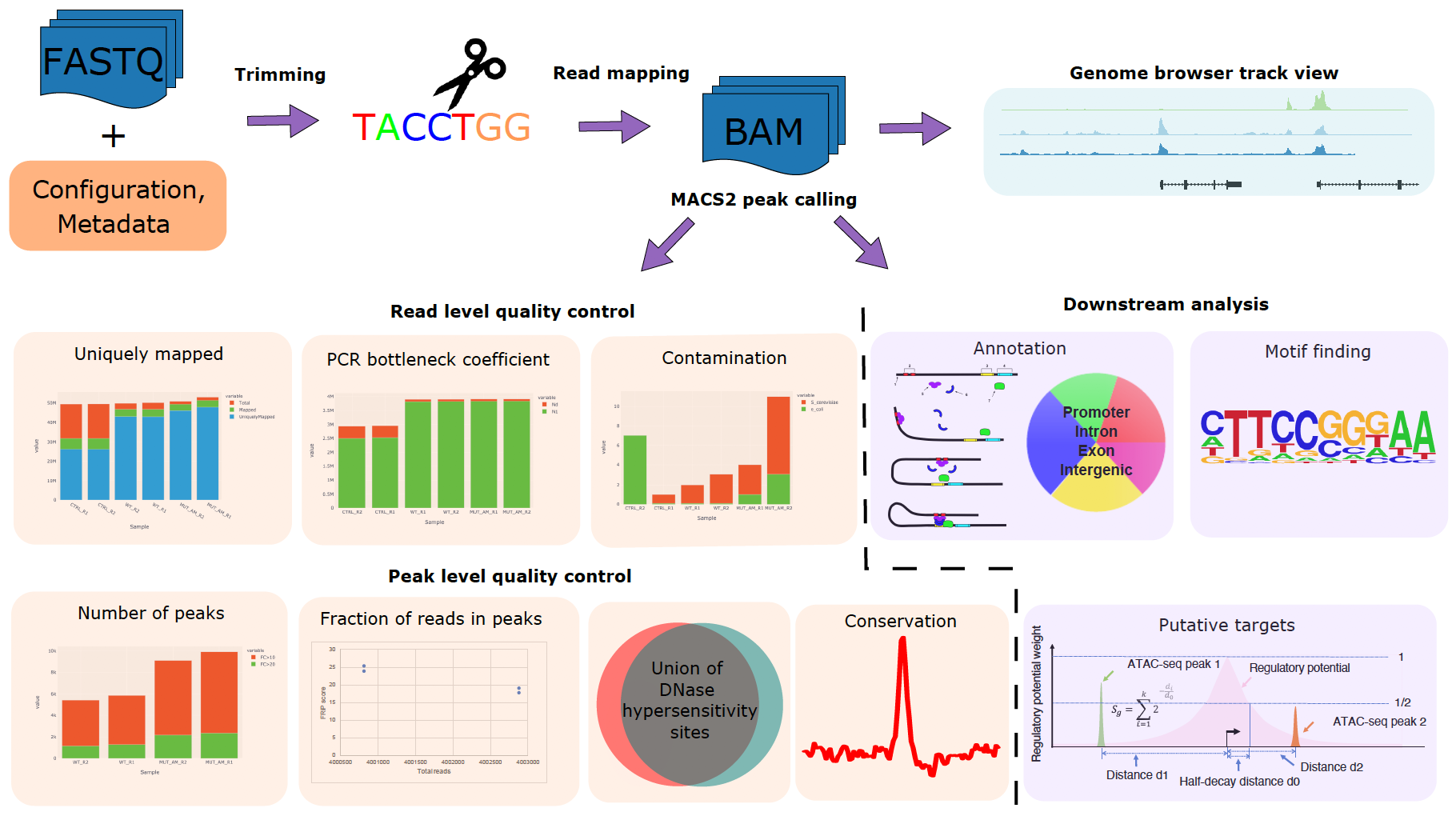
CHIPS: A Snakemake pipeline for quality control and reproducible processing of chromatin profiling data | DNA confesses Data speak

FASTQ Quality Assurance tools 2014 - Bioinformatics Team (BioITeam) at the University of Texas - UT Austin Wikis
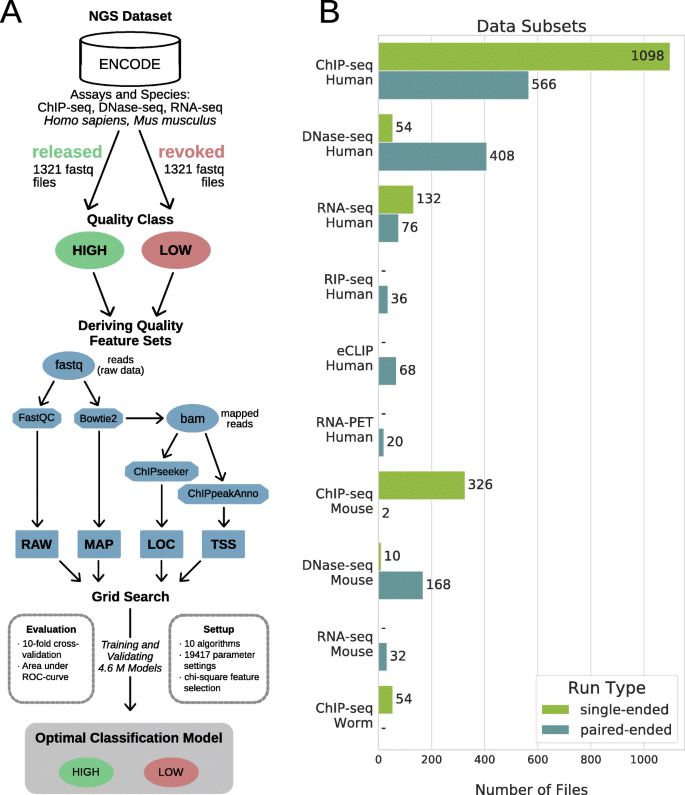
seqQscorer: automated quality control of next-generation sequencing data using machine learning | Genome Biology | Full Text

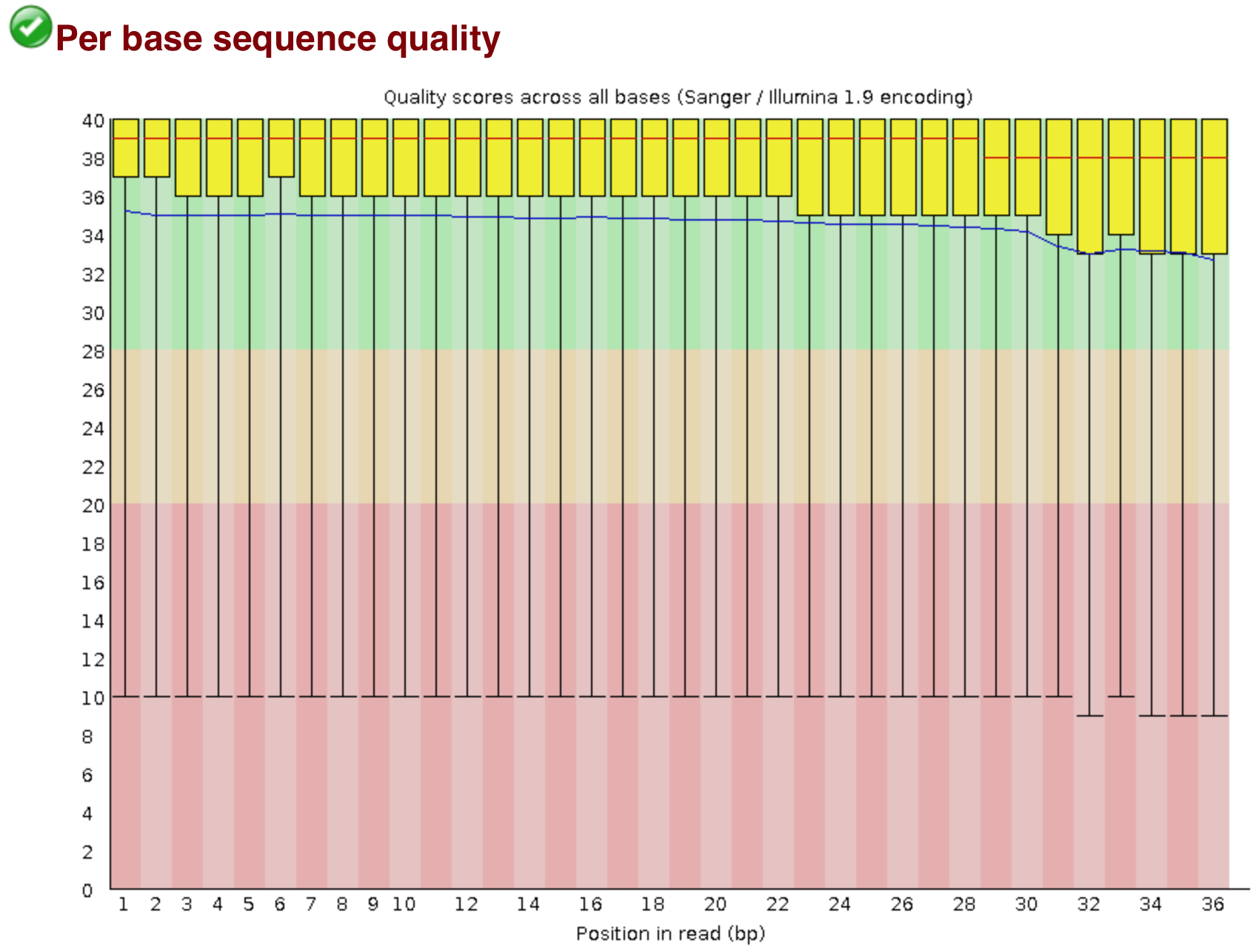
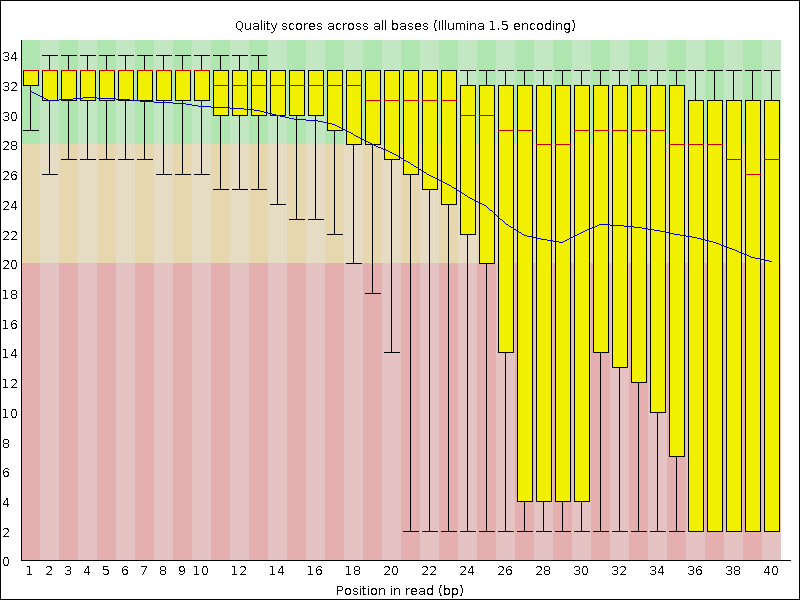
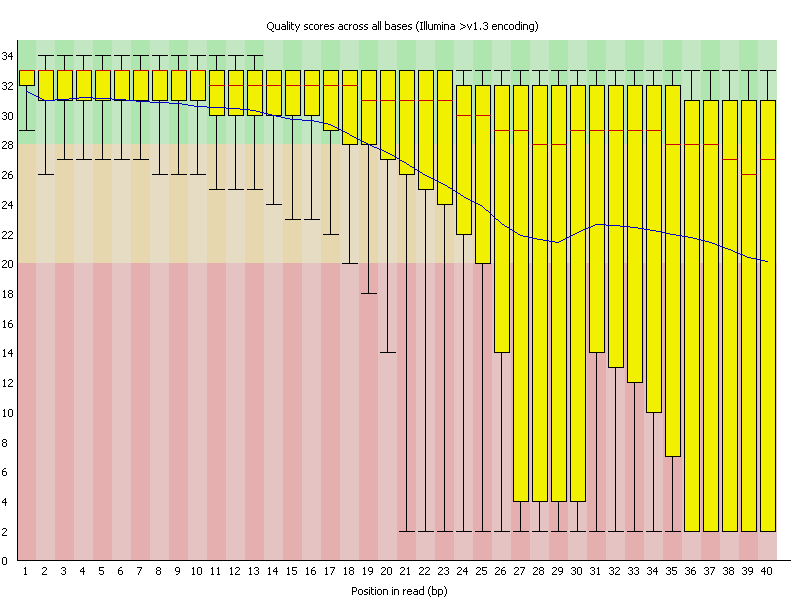
Quality control measures. (a) Per base sequence quality whisker plot:... | Download Scientific Diagram
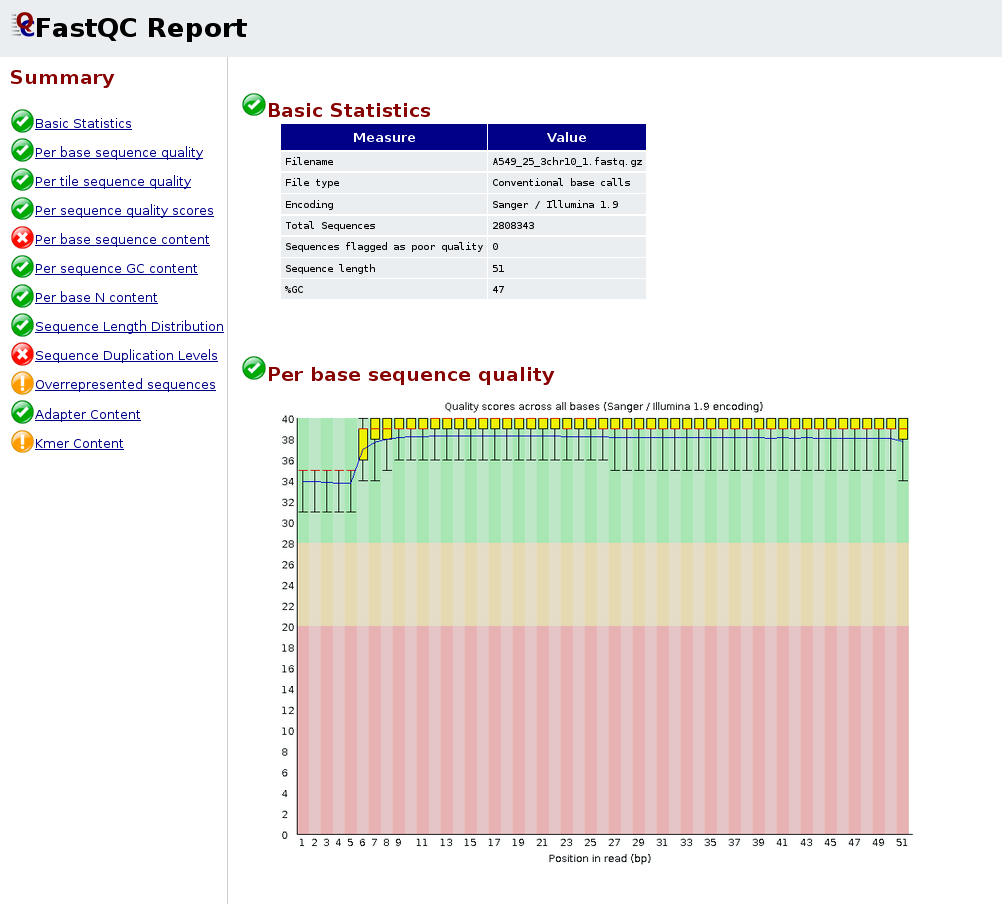
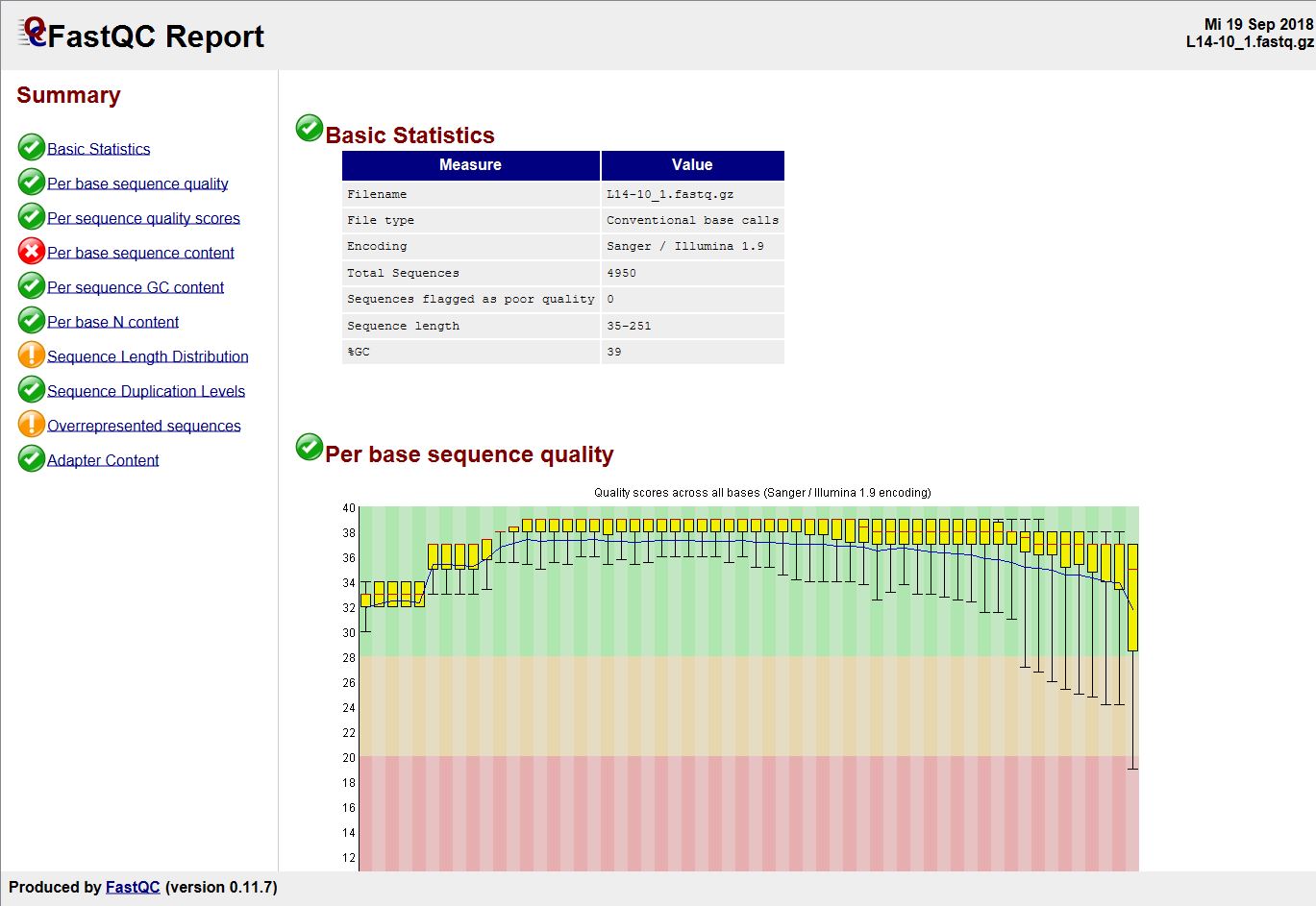
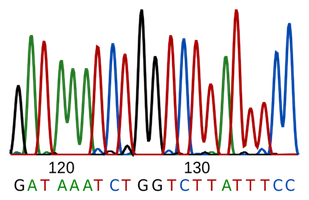
High Throughput Sequence Data Quality Control, Part. 1: FastQC | by ShortLong-Seq Bioinformatics | Medium